Georeactor Blog
RSS FeedEye ML - Training Eyegazer model
For six weeks this summer, I was concerned about a nevus/mole that an optometrist noticed near my optic nerve. It was detected using "fundus photography" which scans the back of the eye. Sample image:
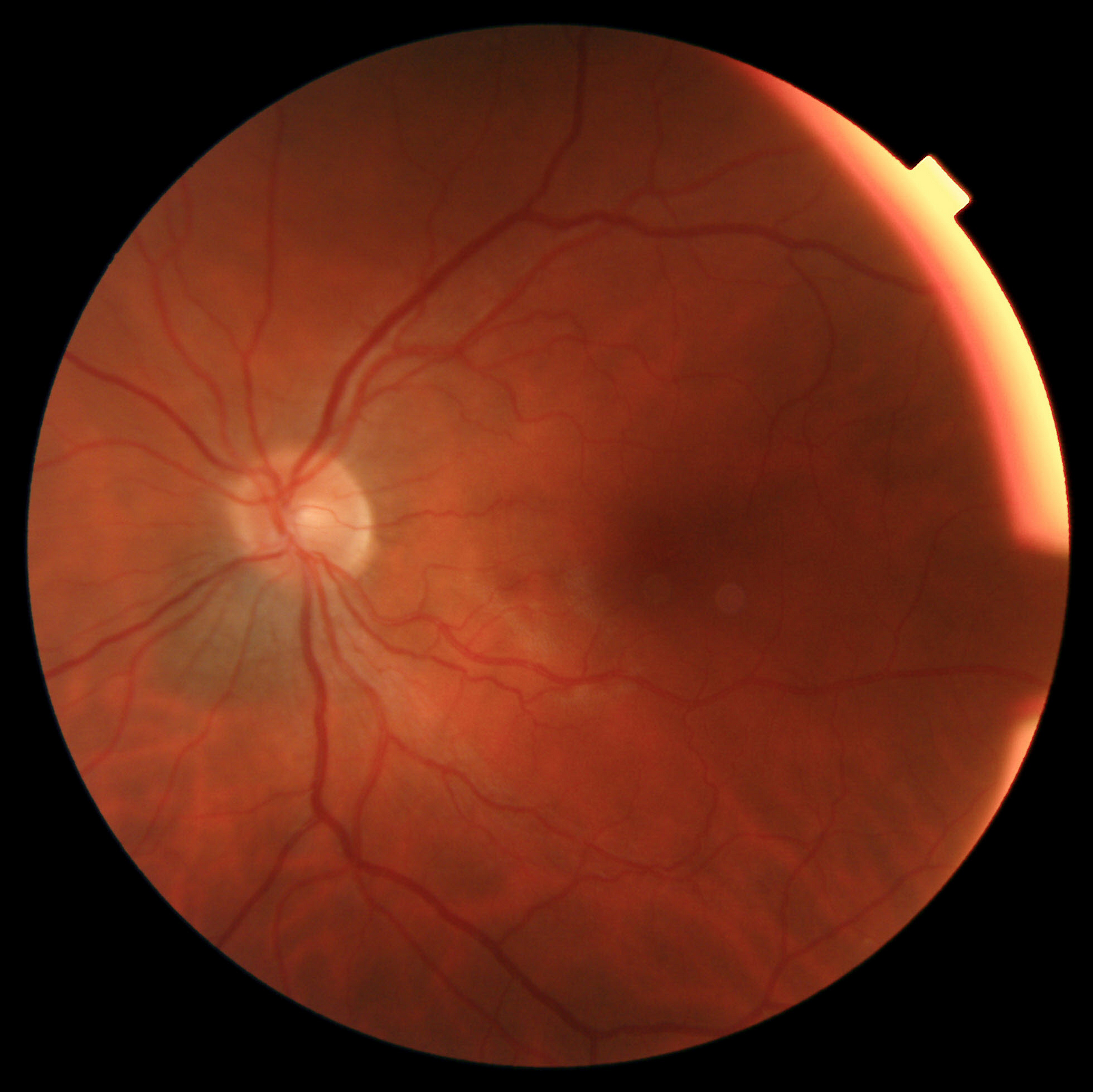
As previously discussed, there are some fundus image competitions on Kaggle. These datasets are generally about detecting diabetic retinopathy, a much more common condition which is under-treated around the world. The Kaggle top score is a 93.5.
As part of my mental process, I wanted to train an eye image model. My original idea was to continue working with IDEFICS or one of the new multimodal LLaMa models.
IDEFICS examples almost always use chat / prompts and not fine-tuning. It seemed unlikely that a general image dataset would have enough eye images for a few-shot prompt to be accurate.
There is one IDEFICS caption fine-tuning example. Full fine-tune and not an adapter. I want to try this, but the given notebook uses a huge dataset which takes a longggg time to load.
LLaMA is a text model, so I'd need to explore community models for a multimodal option. This team did good work with LLaMA v1, but the new multimodal example isn't documented yet: https://github.com/Alpha-VLLM/LLaMA2-Accessory
This took me back to the drawing board for code samples. The best LoRA image classification adapter may be in the HuggingFace PEFT docs. Note this does not support IDEFICS and uses the Google visual transformer (ViT) series of models.
I did a little work ahead of time to reorganize the Kaggle / APTOS 2019 dataset from one big zip into the HuggingFace datasets library format: https://colab.research.google.com/drive/16wyLp6EbVIbA6GvN66itvZHgZFAskV3p?usp=sharing
Fine-tuning run 1: not bad
I use the base-ViT model from the LoRA tutorial. It takes ~20 minutes to train for 3 epochs. A V100 GPU can accept 128 images per batch. Accuracy got up to 76%.
The datasets library loads the label/diagnosis column as a float. I needed to mark it as the special ClassLabel type. Then add label2id and id2label (this is silly when the class conversion is 1='1.0').
Late in the process I realize that these labels are more suited for regression (classification is level 0–4 retinopathy).
The loss does get reduced during training, so it looked right. I was a little concerned with maintaining a consistent copy of the image augmentation and processing pipeline (is the normalization info etc. needed to make consistent predictions?). After paying more attention to the code, it's loading AutoImageProcessor from the original model, so the normalization etc. is constant and carried over from Google's (ImageNet?) pretraining.
Fine-tuning run 2: the shortcut
I upgraded to a larger ViT model, which necessitates a smaller batch size (16) on a V100 GPU. With more batches and more evaluation steps, I noticed this run also achieved 77–78% accuracy, but then fell and stayed constant at 53%.
What happened? I assumed that this was the prevalence of the most common class, and the model was making a shortcut instead of learning traits. In truth it was a little more complicated (about 49% are at 0 and 5% at the maximum level, add them together).
Similar issues may have happened in the earlier model training, just with less visibility.
Fine-tuning run 3: binary
I reformatted the problem to divide between 0 (no diabetic retinopathy) and all other classes. Luckily this is roughly a 50–50 split in training data.
Early runs showed high accuracy, so I changed to two training epochs. It was not possible to use the test dataset for evaluation, because it was intended for Kaggle grading and did not come with labels. It continues to use 10% held-out.
I got some better and worse runs, ultimately getting accuracy in the 90% range. I was satisfied with this as a model and uploaded the LoRA weights as "eyegazer-vit-binary". Note that because LoRA is an adapter, the PeftModel is needed to join it to the original Google ViT Large model.
After working on settings, I also got image classifier inference working on Gradio, though this should be considered only a demo and only for APTOS images.
GPT-4V
Alright, I finally caved and paid money for GPT-4. The ChatGPT interface was reluctant to say anything which could be construed as medical, but we ultimately talked about what to look for in a fundus image, and I shared my own. The model was unable to describe where the nevus was within the image, just mentioning a darker region of the image. Medical industry people are the most likely to need a more customized / controlled / specialized architecture.
Future run: regression
I also chatted with GPT-4 about turning the classifier into a regression problem. There would need to be some work with the interface (setting num_labels=1, possibly adjusting class labels to numbers) or editing the final layer of the network.
Any type of regression model (image or text) is under-documented on HuggingFace. I'd like to either use AutoKeras (where I previously worked on an image regression example doc) or build a training notebook.
Side note on T-cell engagers
For most of the Eye Scare, I assumed that after monitoring, the end result would be radiation. Ocular oncology reccs on YouTube clued me into a newer treatment which got FDA approval in 2022 - I keep forgetting the name but Wiki calls them "Bi-specific T-cell engagers (BiTEs)" and they're a manufactured antibody.
Video Brief for Patients, June 2023
This is not medical advice, and I am not a doctor. The antibody is a puzzle piece / adapter which binds T-cells from the immune system to melanoma cells. Patients need to take a genetic test ahead of treatment to check if they're part of the 50% of the population with matching T-cells (the % here is a little tricky because like other melanomas, ocular melanoma does not appear equally across all populations). What I heard was that in trials this generally did not make the melanoma go away, but the experimental group had noticeably better survival rates than patients where doctors continued usual treatments. Side effects are somewhat common and unpleasant, so it's administered by IV in a hospital setting over multiple sessions.